Welcome to the world of octonary complexes
19th February 2024
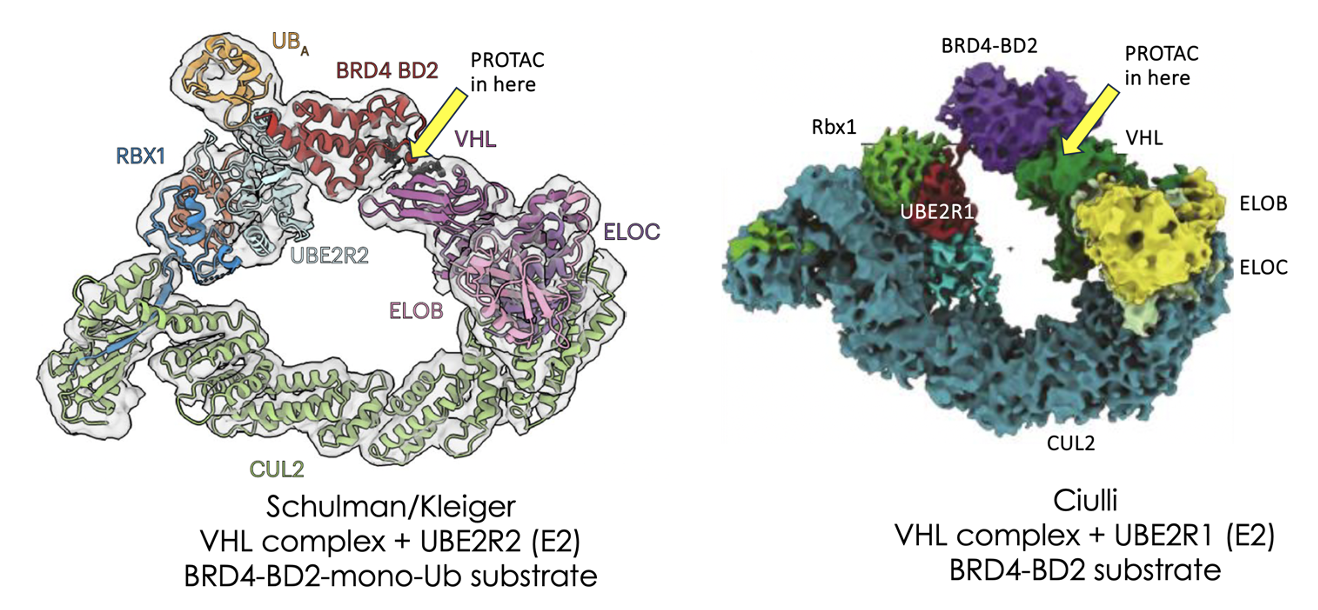
Much hard work is going into the prediction of ternary complex structures between E3 ligases, their substrates and small molecule PROTACs or glues which bridge the two. This is for good reason as understanding this interface is likely to lead to more stable ternary complexes which in turn can give improved degradation.
But we shouldn’t forget, this is not the whole story: the PROTAC or glue is not the “degrader” itself, it simply manages an introduction, helpfully trying to persuade two proteins to get together - a matchmaker hoping to encourage some chemistry between the partners. The real business only happens when complexation allows enough ubiquitins to get transferred from the ubiquitylation complex (E2+E3+accessories) to the substrate in order that it can be recognised by the proteasome (which is the real “degrader” here of course!).
Now, as with so many things in life and business, a successful introduction doesn’t always lead to a productive relationship so we must look beyond the ternary complex to the wider ubiquitylation system which is exactly what a recent publication from the Schulman/Kreiger groups and a preprint manuscript from Alessio Ciulli and his team now start to do for VHL-dependent degradation.
Using CryoEM, which has rapidly become the structural biology tool of choice for large complexes, they elegantly start to show what is going on in the full OCTONARY (8-membered) complex.
Many previous genetic knockout and mutation studies have shown that to get PROTAC-induced ubiquitylation, all 8 members of the complex are required.
For completeness, let’s list the proteins used in the current work along with their role in the ubiquitylation complex:
1) UBE2R [or other E2 ligase which donates ubiquitin to the substrate]; 2) Rbx1 [adaptor], Cullin2 [flexible scaffold at heart of E3 complex – crucially activated by addition of NEDD8 to its flexible WHD domain]; 4) Elongin C [adaptor]; 5) Elongin B [adaptor]; 6) VHL [E3 ligase substrate receptor]; 7) MZ1 [VHL-dependent PROTAC] and 8) BRD4-BD2 [substrate to be ubiquitylated].
As we interpret the information from the structures there’s a few caveats to remember which I’ve summarised at the end of this piece* which you may want to bear in mind.
At the heart of the degrading complexes is cullin4 (cullin2 for the related CRBN-based systems) which elegantly allows the substrate (ie ubiquitin acceptor) to be presented via complex members bound at its N-terminal whilst the E2 (ie ubiquitin donor) is presented via the cullin C-terminal region. The flexibility of the cullin allows these N & C-terminal-bound partners to be brought into close proximity to form a complex giving a perfect circle of ubiquitylating function (see pictures above) and also offers a level of control in the activation state of the complex, an important factor to tune the process. The atomic level detail seen allows some of the key enabling interactions to be mapped.
A series of elegant quench flow kinetic experiments go on to show that the ubiquitin transfer is incredibly fast. Although the E2 ligase can slowly ubiquitylate its substrate alone, when bound to its E3 partner, UBE2R2 becomes one of a small group of enzymes that are considered "at or near catalytic perfection” with its rate limited only by diffusion of the substrate into the active site. This extreme catalytic efficiency is perhaps an evolutionary result due the critical role that ubiquitylation plays in intracellular protein homeostasis. It also makes ubiquitylation an ideal biological process to hijack by small molecule drugs. Much effort is underway to investigate other drug-induced proximity phenomena beyond E3 ligases and it remains to be seen which subset will most closely match PROTAC-like efficiency.
In vitro ubiquitination studies also showed some trends correlating with the octonary complex structure. A total of 8 lysines were ubiquitylated on BRD4-BD2 in the presence of PROTAC MZ1 with all 8 clustering on the side of BRD4 facing Rbx1/E2 suggesting a clear geometric preference for the E2 to donate ubiquitin liberally across a “zone of ubiquitylation”.
Of note, MZ1 is known to selectively degrade BRD4 over related targets BRD2 & BRD3 but when looking at the geometry of the available lysines on BRD2 & 3, whilst slightly different to BRD4, they were largely similar suggesting the apparent selective degradation seen with MZ1 may come mainly from the differential stability of the respective ternary complex interfaces rather than the “intrinsic accessible lysine” landscape on the substrate. Remember, there are a range of parameters which determine PROTAC or glue efficiency which sometimes can't be optimised in isolation.
Looking at ubiquitylation at shorter timepoints, some E2s showed poor specificity in their ubiquitylation pattern though UBE2R1 showed a preference to react with a single lysine, K456. This is interesting as the cryo structure shows K456 appears to be well aligned towards the Ub donor site on the E2. Smoking gun? Well, the cryo structure was far from perfect with several poorly resolved regions and more complete structures may confirm this, but it points to a plausible hypothesis for the atomic level explanation of how ubiquitins get transferred to substrates which is after all what the PROTAC is there to facilitate in the first place.
We should remember though that, despite the elegance of this work, it largely focuses on PROTAC-dependent degrading complexes containing BRD4 as substrate. As those of you familiar with the field will know, BRD4 is studied a lot as it seems to be particularly well suited to PROTAC-induced degradation and indeed there are some degrading approaches which seem to work well for BRD4 and almost nothing else. How easily these conclusions can be generalised to other substrates remains to be seen. Also of course, currently most TPD activity uses not VHL but CRBN-based systems. As degradation here is mediated by only septenary (7-member) and not octonary (8-member) complexes, they may be simpler to model :-)
These cryo structures offer us an intriguing snapshot into what is undoubtably a complex process with different lysines ubiquitylated at different rates in a likely ensemble of dynamic complexes but the concept of E2-accessible lysines or a zone of ubiquitylation may be useful to assess the degradability of some substrates (as has been suggested by several groups see eg here).
Modelling intact octonary complexes at the atomic level may be a challenge for even the most ambitious molecular dynamics or other computational simulations though it’s likely that some assumptions and constraints can be applied to reduce the number of dimensions of variability somewhat without detracting from the physiologic relevance too much. Figuring out which shortcuts can be safely taken may be key to getting the best use out of this kind of information for identifying and optimising new degrading drugs. This insight can undoubtedly help us design better drugs...when used thoughtfully.
*Some important caveats to remember:
There’s a huge amount of great work in these papers and I’ve only picked out a few areas to discuss here but there’s some crucial details we need to remember. E2 ligases which donate the ubiquitin come in two main flavours – “priming” E2s which add the first Ub to the substrate (often UBE2G or UBE2D family) as well as “extending” E2s which then propagate the Ub chains via K48 linkages to give the familiar poly-Ub decoration which is the signal for the proteasome to recognise and degrade the substrate. The “extending” E2 is often and as in the current work, a UBE2R family member E2. Different E2 complexes are likely to give different ubiquitylation.
Also, the current cryo structures try to mimic ubiquitylation transition states which are necessarily ephemeral and tough to observe so a few tricks in terms of pre-linking the substrate and E2 are needed in order to get complexes observable in cryo-EM. (The Schulman paper uses monoubiquitylated BRD4 covalently bound to an E2-Ub system [ie mimics chain extension] while the Ciulli team bind the E2-Ub to unubiquitylated BRD4 via a bismaleimide linkage [ie mimicking chain priming])
In some cases, Cryo EM reveals a complex ensemble of species which, n the case of the Ciulli work, when deconvoluted showed several states including an “open” state where the BRD4 was not engaged with the Rbx1 terminus as well as mixed, dynamic (ie lower resolution) states suggesting a “closed” conformation where the VHL and Rbx1/E2 regions were in close contact. This again nicely shows the degree of control that the overall geometry including he Cullin conformation and NEDDylation state can have in the activity of the complex.
Finally, due to the large size of the overall octonary complex and presence of flexible regions, several areas are poorly resolved or missing including the NEDD8 & other areas which may lead to limitations in interpretation.
Bearing in mind these caveats, the structures offer a fascinating insight into the ubiquitin transfer process however!
We need your consent to load the translations
We use a third-party service to translate the website content that may collect data about your activity. Please review the details in the privacy policy and accept the service to view the translations.

