Not all E3 ligases are born the same. Are you feeling lucky?
23rd October 2024
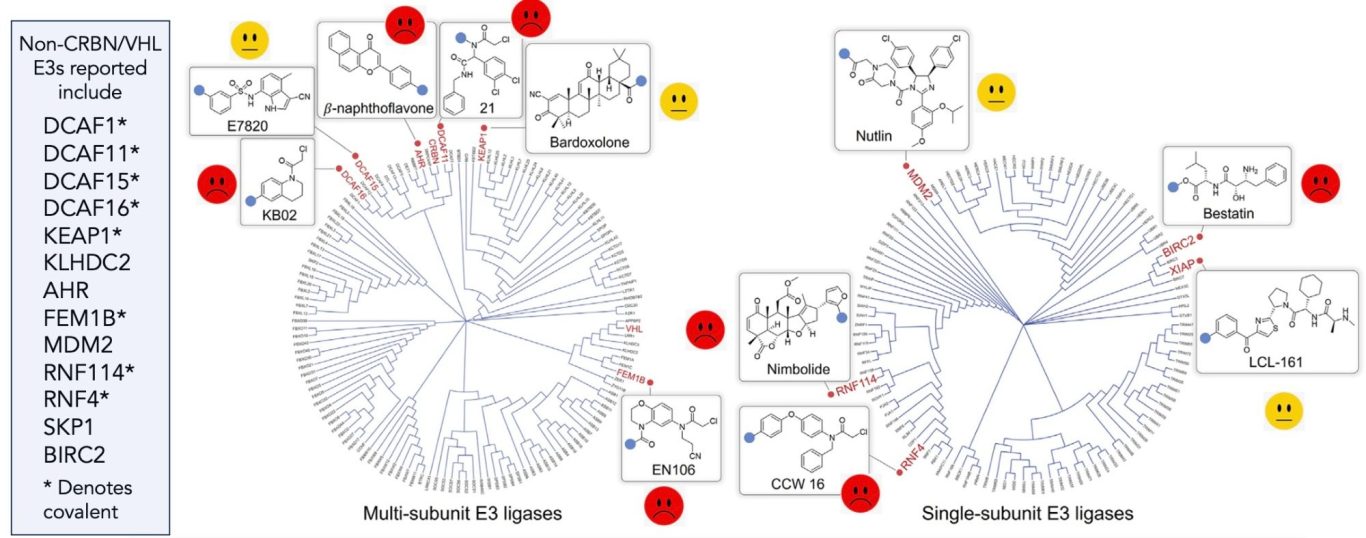
“There are over 600 E3 ligases…” is a phrase you’ll see used all the time in TPD presentations and publications. So why is it we always default to using the same one or two in cereblon (CRBN) and von Hippel Lindau (VHL)?
There may be a few factors at play here which make CRBN & VHL particularly good at being repurposed to degrade your substrate of choice while other E3 ligases – well, they just don’t want to play ball and are just happy degrading their physiological substrate and probably not much else.
Whether your chosen degrading strategy is a PROTAC or a glue, you’ve probably spent a lot of time using CRBN-based degraders but why is this specific E3 ligase so good at degrading such a wide range of substrates? The elegant work of Christina Woo gives us a big hint. Christina and team showed that CRBN appears to recognise a glutarimide/succinimide motif (cf the key pharmacophore always seen in lenalidomide, pomalidomide and almost all other CRBN ligands) which can appear in proteins after a decomposition pathway involving intramolecular cyclization of glutamine or asparagine residues to leave a CRBN-friendly C-terminal imide degron.
This of course all makes sense. As proteins age, they start to acquire often undesirable post-translational modifications (ie damage) through eg oxidative stress, misfolding or unwanted cleavages. A damaged protein may start to misbehave in several ways including losing its normal physiologic function or, more concerningly, acquiring a new function eg aggregating or binding to things it shouldn’t. At this point, the cell has a vested interest in identifying these damaged proteins and getting rid of them before they cause problems so having an E3 ligase which goes round looking for truncated proteins bearing cyclic imide motifs is evolutionarily, a good thing.
Hence, CRBN may be a “housekeeping” E3 ligase that is very good at binding to anything with an imide pharmacophore then ubiquitylating it and consigning it to a rapid demise in the proteasome. As CRBN is a housekeeper, it needs a broad substrate scope, perhaps facilitated by a certain amount of flexibility and plasticity in its binding surface which allows it to form often highly co-operative glue/PROTAC ternary complexes with many substrates. Indeed, recent studies have identified over 1400 proteins which may be good CRBN substrates.
Are there other housekeeping E3 ligases? Well, maybe. The list of non-CRBN, non-VHL E3s which give efficient degradation of a wide range of substrates is not a long list (slowly getting longer, see for example the expanding KLHDC2 work).
As an aside, it’s worth noting at this point that many new E3s first show their worth in TPD by degrading BRD4. Now that’s ok but BRD4 does not appear to be a typical target. In the same way that CRBN seems well suited to form ternary complexes to degrade things, then BRD4 also appears to be particularly adept at forming complexes and being itself degraded. There are examples of new TPD approaches which are applicable to potently degrade BRD4 and little else so beware if you choose this as a probe substrate. The world doesn’t really need any more BRD4 degraders and you could be misled into thinking your shiny new TPD approach is more generally applicable than it really is.
Back with the new E3s, a number of DCAF (DDB1 Cullin4 associated factor) family members are now starting to come up more regularly as useful E3s, albeit some of the ligands used are pretty reactive. Let’s not forget of course that CRBN is actually a DCAF family member and it’s only a quirk of naming that it wasn’t the original DCAF1 before the family got better characterised. It's also perhaps no coincidence that most of the pharmacophores which bind to DCAFs are electrophilic in nature. Many aged and damaged proteins can become electrophilic, often through oxidative processes but strong electrophilicity in the cell is generally a very bad thing given that cells are mainly bags of nucleophiles (OH, SH, nitrogen bases etc). An out of control electrophile could cause a lot of biochemical chaos so it’s plausible that DCAFs may be a family of housekeeping E3s which have evolved to go round looking for electrophilic proteins and ubiquitylating and eliminating them. Make a weakly electrophilic warhead and incorporate it into your PROTAC/glue to bind to your protein of interest and there’s a good chance you’ll attract the attention of one or more housekeeping DCAFs to neatly remove your desired protein.
But, what about VHL? Could that be a housekeeping ligase too? VHL has exquisite selectivity for binding to the prolyl hydroxylated form of its substrate hypoxia-inducible factor (HIF) which is then degraded as a critical step in the cellular oxygen sensing pathway. The exact range of proteins which could be VHL substrates via acquisition of hydroxyproline marks remains an area of some uncertainty but the range may not be large. So why is VHL apparently repurposed so easily to degrade new substrates (via PROTACs at least, glues look to be much rarer)?
Due to the criticality of oxygen sensing in the cell, VHL has likely evolved to be very, very good at enabling ultra-rapid ubiquitylation of hydroxylated HIF. With an E3 which is just THAT GOOD, perhaps it can still efficiently mediate ubiquitylation of other substrates. Even if the ubiquitylation rate of a new substrate is only a fraction of that seen with HIF, it may still be more than fast enough that your VHL PROTAC becomes a good degrader of your substrate of choice. So maybe VHL isn’t truly a housekeeper, it’s just an amazingly efficient and fast-acting E3 where even relatively slow PROTAC-mediated ubiquitin transfer to non-HIF substrates is still really rather good.
Perhaps then, outside a small set of housekeepers and ultrafast E3s, maybe the rest are just plain old hardworking E3s which do a great job processing their defined set of natural substrates in the cell but really don’t want to be persuaded to degrade much else so won’t be much use for therapeutic TPD applications? We’ll see.
But what if you really want to find an alternative E3 instead of using a CRBN or VHL-based degrader, what are your options?
- You could select a ligase with tissue-selective expression then go and find ligands and turn them into PROTACs/glues etc. I wouldn’t recommend this as the ligandability of many E3s is low and even if you do find ligands, you’re unlikely to select a ligase which is readily repurposable so you could spend an awful lot of time and resource going down this route. Many people have tried and success stories are conspicuous by their absence - though it’s not impossible that someone has cracked it and is understandably holding the information close to their chest (though you’d expect to see patent applications starting to filter out with clues).
- Alternatively, to reduce the low ligandability risk, you could select a ligase predicted to be highly druggable then use eg DEL screening to find ligands then turn them into PROTACs, glues etc. Again, I may not recommend this for the same reasons as above. Even if you find good ligands, turning them into degraders of targets you care about is tough as the E3 may just not want to be repurposed. Again, many people have tried but the success stories are not generally in the literature.
- Finally, you could identify E3 ligases with wide substrate scopes or ultrafast rates of ubiquitylation. This could be more useful as you’ll be more likely to find more “CRBN or VHL-like” ligases which are more likely to accept a range of new substrates. Sure, you still need to find ligands to the ligase but at least when you do, there’s a higher chance that the PROTACs or glues you derive from them will actually be useful to degrade new substrates that you’re interested in.
Of course, instead of being forced to pick one ligase out of 600, you could take a ligase agnostic approach and run a cellular phenotypic screen to look for degraders of your desired substrate which could use any of the 600. This has been a successful glue discovery strategy and more people are trying this for other degraders now but in some cases, even in an unbiased screen, all roads lead to CRBN! The path of least resistance is very attractive sometimes.
Finally, if you feel that neither CRBN-based degraders or VHL-based degraders are quite good enough, then why not make degraders which use both of them? That approach certainly works though the molecules start to wave farewell to oral delivery potential but you can certainly see advantages in terms of broader cell scope or lower risks of tumors developing resistance.
In conclusion, when it comes to TPD therapeutics, some ligases just work better than others and we should continue to take full advantage of this. There is still much more value to be gained from CRBN, VHL and an expanding but small set of other ligases. Then there’s the rest of the 600. There could be some gems in there waiting to be discovered but this vein has been mined a lot over the last 10 years so, are you feeling lucky?
We need your consent to load the translations
We use a third-party service to translate the website content that may collect data about your activity. Please review the details in the privacy policy and accept the service to view the translations.

